This blog post is based on the net surgery example provided by Caffe. It takes the concept and expands it to a working example to produce pixel-wise output images, generating output in ~2 seconds (simple approach) or ~35 seconds (advanced approach) for a 2,000 x 2,000 image, an improvement from the ~15 hours of a naive pixel wise approach.
Introduction
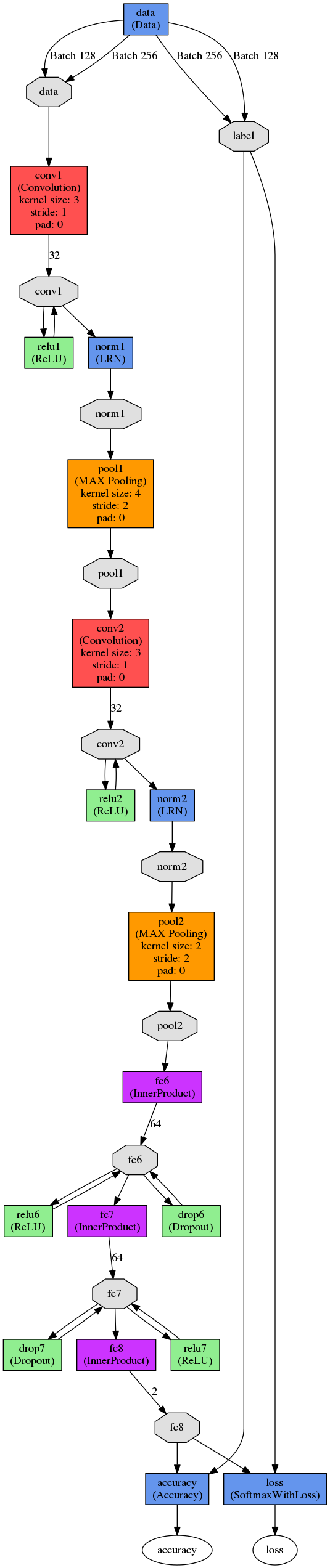
The net surgery example discusses how to take a patch level classifier and convert it to a fully convolutional network. The underlying concept is that convolutions are agnostic to the input size, but that the fully connected layers need to be modified such that they can be computed in a convolutional fashion. This is done by modifying both the deploy file and the associated Caffe model.
Here we present two approaches. A fast (simple) approach which runs the classifier once across the image and uses it to interpolate the necessary pixel values to generate a full size image pixel wise image. The second (advanced) approach runs the DL classifier multiple times at different displacement offsets to generate a very accurate image. While the computational cost is higher (35 seconds vs 1.5 seconds), it is still a far cry from the original 15 hours it would take to perform the same operations pixel-wise.
Architecture Constraints
The network architecture used for this process cannot have any padding at any of the levels. This should make some intuitive sense. As we’re operating on the whole image at a single time, the only padding which would take place are at the absolute edges. When operating at the patch level, the padding occurs on the edges of every patch, providing either 0’s or mirrored pixels (in the case of caffe the padding style is 0s).
The easiest way to check is by using grep:
- axj232@ccipd-tesla-001:~/caffe/examples/ajtemp$ cat deploy.prototxt | grep pad
- axj232@ccipd-tesla-001:~/caffe/examples/ajtemp$
If there is no output, then no padding variables are set, and we should be good to go!
Here is an example of the network we’re using, note the lack of padding (click for large version):
Conversion
We use the same process as described in the net surgery example, and explicitly state each of the steps.
1. Copy the deploy.prototxt to a deploy_full.prototxt
2. Identify all InnerProduct layers, in this case, fc6, fc7 fc8:
- $ grep -B 1 Inner deploy.prototxt
- name: "fc6"
- type: "InnerProduct"
- --
- name: "fc7"
- type: "InnerProduct"
- --
- name: "fc8"
- type: "InnerProduct"
Replace all instances of the layer name with an appended “-conv”, e.g., fc6 -> fc6-conv, fc7 -> fc7-conv, etc. This prevents Caffe from accidentally loading the old values from the model file and more accurately represents the layer type (remember, we’re converting them from fully connected to convolutional).
3. Replace “InnerProduct” with “Convolution”
4. Replace “inner_product_param” with “convolution_param”
5. Add “kernel_size: 1″ to convolutional_param for all these layers except for the first layer we’re changing (i.e., fc6-conv).
6. Fc6-conv is a bit trickier to figure out, we need to know the exact size of the kernel which comes in. This is trivially done if we look at the training log from the patch level classifier, as it states the input and output sizes of each layer as it scaffolds them:
- I1220 17:28:27.392643 2183 net.cpp:106] Creating Layer pool2
- I1220 17:28:27.392701 2183 net.cpp:454] pool2 pool2
- I1220 17:28:27.392806 2183 net.cpp:150] Setting up pool2
- ---> I1220 17:28:27.392823 2183 net.cpp:157] Top shape: 128 32 6 6(147456)
- I1220 17:28:27.392837 2183 net.cpp:165] Memory required for data: 56689152
- I1220 17:28:27.392848 2183 layer_factory.hpp:76] Creating layer fc6
- I1220 17:28:27.392873 2183 net.cpp:106] Creating Layer fc6
Here we can that the top shape is 128 x 32 x 6 x 6. 128 is our batch size, 32 is the num_output from the previous convolution later, leaving us with a 6 x6 kernel. As a result, we set fc6-conv “kernel_size: 6”.
7. We need to specify the exact input size of the image we’ll be using, otherwise Caffe will resize (and possibly change the aspect ratio) the size of our input image to the specification size. In this case, we had a 32 x 32 patch classifier, and we want to use it on 2032 x 2032 images, so we modify the deploy file accordingly:
| Old | New |
| input_shape { dim: 1 dim: 3 dim: 32 dim: 32 } |
input_shape { dim: 1 dim: 3 dim: 2032 dim: 2032 } |
Note that our actual input image is 2000 x 2000, except we want to be able to compute the edges, so during output generation time we’ll mirror the edges by exactly half the patch size. In this case, that comes to +16 pixels on each size.
8. Depending on the method used to make the deploy file (such as NVIDIA Digits) , we may need to add the bottom softmax layer to get a probability distribution, this is done by simply adding to the bottom of the file:
- layer {
- name: "prob"
- type: "Softmax"
- bottom: "fc8-conv"
- top: "prob"
- }
With that information, we can use convert-patch-classifier-to-full-convolutional.py to make a new caffe model for us. Here we can see the old and new deploy models.
Output Generation (Simple)
Using our new network, we simply need to feed the image into the classifier and get the output. Note that the size of the image is 2000 x 2000, we specified a padded size of 2032 x 2023 above in the deploy text. The padding occurs in cell [6]. These sizes need to be the same!
We can see the code in action here.
Here we note two thing, first off, we’ve produced our input image in ~2 seconds. Not bad!
Secondly, we can see that the intermediate output image (im_out) is actually of size 501 x 501 and not the 2000 x 2000 we expected. This makes some sense, since at each layer we reduce the data size through convolution and pooling. The python script takes these values and interpolates them correctly back to the larger image.
Depending on your image size and network characteristics, this is a critical step. We define what we call the “image_ratio”, which is the ratio from the larger image to the smaller image, in this case 2000 x 501 = 3.992.
Since this image ratio is not an integer (and can be “worse” such as 3.5), it has a compounding effect on the pixel location in im_out versus im_out_final. For example, if we have an output image of 500 x 500, the image ratio would put the 500th pixel at 500 * 3.99 = 1995. When, we know for a fact that it comes from an image of 2000 x 2000. We can see that as the images get larger the image ratio introduces more and more error, in this case, the pixel is incorrectly located 5 pixels away from its “real” location. This is why the interpolation is so important!
Below we can see the result from a nuclei segmentation task, as generated by this new efficient code (left) versus the pixel-wise naive approach (right). Looking nearly identical, but with 99.996% less computation time!
Output Generation (Advanced)
At this point, we already have a very rapid output generation approach, but we’re not really computing the network at each pixel. In effect we’re computed every image_ratio pixel and interpolating the ones in between. While this is more than sufficient in most cases, is it possible to compute the pixels in between and interlace them in an efficient manner? It certainly is 🙂
If we know our image_ratio, we can displace the original image inside of this “receptive field“, compute output at each displacement, and then merge them together. More explicitly, compute row/column displacements of image_ratio
. In this case, that’s
operations. If each operation took ~2 seconds before, we’d expect to get them all done in ~30 seconds. Still pretty impressive!
At a high level the results look the same between the two methods, but if we zoom in on a 288 x 271 region, we can see some striking differences:
Simple Approach
Advanced Approach
Where the left image shows the overlay of the right image on this original image
We can see the code in action here.
That pretty much sums it up! The entire code base is available here.




Hi,
I tried to replicate your experiments but whenever I try to load the full convolutional network along with the caffe model (in convert-patch-classifier-to-full-convolutional.py) the program crashes saying: Cannot copy param 0 weights from layer ‘conv1’; shape mismatch. Source param shape is 96 3 11 11 (34848); target param shape is 32 3 3 3 (864). Any idea what can be wrong? Thanks!
It seems that the deploy file that you’re using and the modified fully convolutional deploy file don’t specify the same network. In this case, one layer is specified as having 96 x 3 x 11 x 11 inputs (with 34,848 variables) and it is trying to reshape that into a layer with 32 x 3 x 3 x 3 (864 variables). If I were you, i’d go line by line and assure the networks are the same. Hoep that helps!
Hey,
We are trying to tackle a similar problem, but our dataset is very unbalanced – we are trying to detect folds in histological sections (binary pixelwise classification – either a fold or “no fold”) and most of the image is “no fold”. I tried replicating this architecture, but it fails of course because of an unbalanced dataset. So how did you handle this? Did you use any pre-trained weights? Please let me know, maybe on email as well.
i would say that there are 2 approaches, you can either hyper-sample the smaller class so that the training set is somewhat balanced, or hypo-sample the over represented class and then use something like transfer learning to help to greatly reduce the minimum size of the dataset needed. the first approach is similar to the mitosis use case discussed in the paper, where the # of negative patches is significantly larger than the # of pixels associated with a mitosis
Hi Andrew,
Have you ever simulated data for patterns such as tissue folding using DL approaches ? The interesting thing about this type of patterns is that at low resolution texture parameters are quite similar and using affine transformation you can simulate data to cover all possible forms and combined with shifting of intensity levels you can also cover the staining variability.
Using such simulated data and more classical supervised machine learning models we have achieved good performances but was wondering if you guys have been exploring this type of simulations for DeepLearning frameworks ?
Elton
I’ve briefly looked at using simulated (synthetic) data for DL augmentation. Its interesting that most of the augmentations, if they’re rather trivial (lighting augmentation, etc), don’t seem to improve the quality or robustness of our classifiers. Its almost like the first layer of the network becomes completely agnostic to them.
Typically what I’m finding is that DL is very accurate in “easy” situations, so if you consider a nuclei segmentation case, the obvious pixels in the center of the nuclei and the obvious non-nuclei pixels tend to do very well. The biggest challenge is in the transition zone, i.e., right on the boundary of a nucleus. This should make some type of sense as we would consider those points to be closer to the decision boundary in the higher dimensional space, so having more labeled points would allow for a more precise boundary that is more likely to generalize well. Unfortunately, these types of “difficult” cases have been rather hard for us to create in a meaningful way for the types of problems that we’re looking at.
One possibility is using Generative Adversarial networks (GANs), but I would want to ask the question, lets say I asked the GAN to produce synthetic cancers, (a) is it going to produce a biologically plausible cancer (otherwise its likely useless for generalization) and (b) are the cases it generates going to be the “hard” cases that we need to improve the classifier as opposed to the “trivial” examples I mentioned above. All open questions for research as far as I understand