This is an updated version of the previously described workflow on how to load and classify annotations/detections created in QuPath for usage in downstream machine learning workflows. The original post described how to use the Groovy programming language used by QuPath to export annotations/detections as GeoJSON from within QuPath, made use of a Python script to classify them, and lastly used another Groovy script to reimport them. If you are not familiar with QuPath and/or its annotations you should probably read the original post first to provide better context and understanding of the respective workflows, as well as being able to appreciate the more elegant approach taken here. If you are already using the described approach, you should be able to easily modify it to follow this newer approach.
Continue reading Using Paquo to directly interact with QuPath project files for usage in digital pathology machine learningTutorial: Quick Annotator for Tubule Segmentation
The manual labeling of large numbers of objects is a frequent occurrence when training deep learning classifiers in the digital histopathology domain. Often this can become extremely tedious and potentially even insurmountable.
To aid people in this annotation process we have developed and released Quick Annotator (QA), a tool which employs a deep learning backend to simultaneously learn and aid the user in the annotation process. A pre-print explaining this tool in more detail is available [here].

Transferring data FASTER to the GPU With Compression
Utilization of current GPUs is often limited by the ability to get the data onto and off the device quickly. More precisely, this means taking data from the host RAM, transferring it over the PCI-e bus to the GPU RAM is the bottleneck of many deep learning use cases.
Continue reading Transferring data FASTER to the GPU With CompressionDe-array a Tissue Microarray (TMA) using QuPath and Python
This blogpost demonstrats how to use QuPath and Python to de-array a tissue microarray (TMA) for computational analysis and was co-authored by Fan Fan (fxf109@case.edu)
Continue reading De-array a Tissue Microarray (TMA) using QuPath and PythonThe noise in our digital pathology slides
In adding new features to HistoQC , I stumbled upon a very interesting insight that I thought I would take a moment to share. The amount of noise and artifacts in digital pathology (DP) whole slide images (WSI) is far more extensive than I had previously thought.
Continue reading The noise in our digital pathology slidesConverting an existing image into an Openslide compatible format
Many digital pathology tools (e.g., our quality control tool, HistoQC), employ Openslide, a library for reading whole slide images (WSI). Openslide provides a reliable abstraction away from a number of proprietary WSI file-formats, such that a single programmatic interface can be employed to access WSI meta and image data.
Unfortunately, when smaller regions of interest, or new images, are created in tif/png/jpg formats they no longer remain compatible with OpenSlide. This blog post discusses how to take any image and convert it into an OpenSlide compatible WSI, with embedded metadata.
Continue reading Converting an existing image into an Openslide compatible formatExporting and re-importing annotations from QuPath for usage in machine learning
Update-Nov 2020: Code has now been placed in github which enables the reading and writing of compressed geojson files at all stages of the process described below. Compression reduces the file size by approximately 93% : )
QuPath is a digital pathology tool that has become especially popular because it is both easy to use to and supports a large number of different whole slide image (WSI) file formats. QuPath is also able to perform a number of relevant analytical functions with a few mouse clicks. Of interest in this blog post is mentioning that the pathologists we tend to work with are either already familiar with QuPath, or find it easier to learn versus other tools. As a result, QuPath has become a goto tool for us for both the creation, and review of, annotations and outputs created by our algorithms.
Here we introduce a robust method using GeoJSON for exporting annotations (or cell objects) from QuPath, importing them into python as shapely objects, operating upon them, and then re-importing a modified version of them back into QuPath for downstream usage or review. As an example use case we will be looking at computationally identifying lymphocytes in WSIs of melanoma metastases using a deep learning classifier.
Continue reading Exporting and re-importing annotations from QuPath for usage in machine learningComputationally creating a PowerPoint presentation of experimental results using Python
This post is an update of the previous post, which discussed how to create a powerpoint slide desk with results using Matlab. In the last couple of years, we have mostly transitioned to python for our digital pathology image analysis, in particular those tasks which employ deep learning. It thus makes sense to port our tools over as well. In this case, we’ll be looking at building powerpoint slide desks using python.
Let’s look at what we want as our final output:

SDiPath – Introduction to Computational Pathology for Pathologists
Thanks to everyone in Bern for their attendance at our workshop!
Helping to introduce these concepts to our clinical collaborators is incredibly important for advancing our field, so if you’re interested in hosting a workshop, please feel free to reach out!
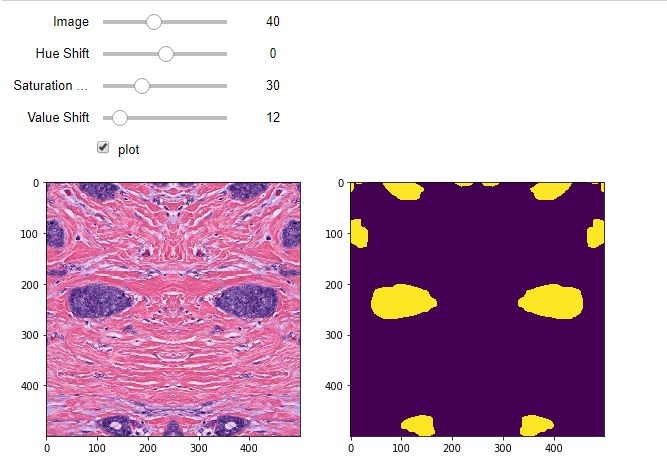
Employing the albumentation library in PyTorch workflows. Bonus: Helper for selecting appropriate values!
This brief blog post sees a modified release of the previous segmentation and classification pipelines. These versions leverage an increasingly popular augmentation library called albumentations.