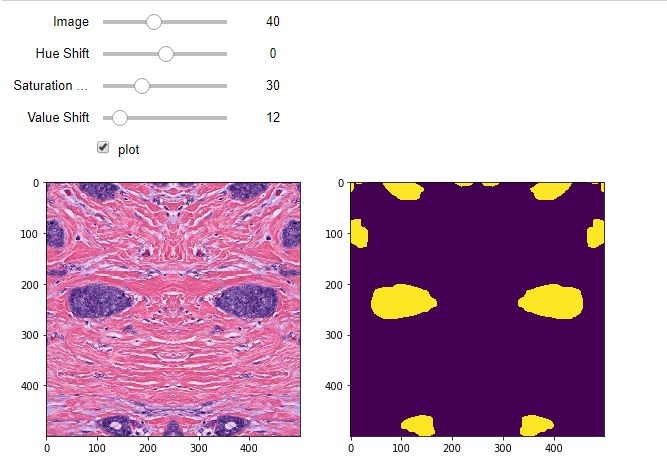
This brief blog post sees a modified release of the previous segmentation and classification pipelines. These versions leverage an increasingly popular augmentation library called albumentations.
Tag Archives: python
Image popups on mouse over in Jupyter Notebooks
Animation below speaks for itself : )
Finally put together a script which makes jupyter notebooks plots interactive, such that when hovering over a scatter point plot, the underlying image displays, see demo + code below:
Code is available here: https://github.com/choosehappy/Snippets/blob/master/interactive_image_popup_on_hover.py
Digital Pathology Segmentation using Pytorch + Unet
In this blog post, we discuss how to train a U-net style deep learning classifier, using Pytorch, for segmenting epithelium versus stroma regions. This post is broken down into 4 components following along other pipeline approaches we’ve discussed in the past:
- Making training/testing databases,
- Training a model,
- Visualizing results in the validation set,
- Generating output.
This model focuses on using solely Python and freely available tools (i.e., no matlab).
This blog post assumes moderate knowledge of convolutional neural networks, depending on the readers background, our JPI paper may be sufficient, or a more thorough resource such as Andrew NG’s deep learning course.
Continue reading Digital Pathology Segmentation using Pytorch + Unet
Using Matlab, Pytables (hdf5) and (a bit of) Pytorch
As we’re testing out for migration to new deep learning frameworks, one of the questions that remained was dataset interoperability. Essentially, we want to be able to create a dataset for training a deep learning framework from as many applications as possible (python, matlab, R, etc), so that our students can use a language that are familiar to them, as well as leverage all of the existing in-house code we have for data manipulation.
Continue reading Using Matlab, Pytables (hdf5) and (a bit of) Pytorch
Real time Data Augmentation using Nvidia Digits + Python Layer
One of the common ways of increasing the size of a training set is to augment the original data with a set of modified patches. These modifications often include (a) rotations, (b) mirroring, (c) lighting adjustment, (d) affine transformations (sheering, etc), (e) magnification modification, (f) addition of noise, etc. This blog post discusses how to do the most trivial modification, rotation, in real-time using a python layer through Nvidia Digits. Given this code, it should be easy to add on other desired augmentations.
Continue reading Real time Data Augmentation using Nvidia Digits + Python Layer
Revised Deep Learning approach using Matlab + Caffe + Python
Our publication “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases” , showed how to use deep learning to address many common digital pathology tasks. Since then, many improvements have been made both in the field and in my implementation of them. In this blog post, I re-address the nuclei segmentation use case using the latest and greatest approaches.
Continue reading Revised Deep Learning approach using Matlab + Caffe + Python
Use Case 7: Lymphoma Sub-Type Classification
This blog posts explains how to train a deep learning lymphoma sub-type classifier in accordance with our paper “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases”.
Continue reading Use Case 7: Lymphoma Sub-Type Classification
Use Case 5: Mitosis Detection
This blog posts explains how to train a deep learning mitosis detector in accordance with our paper “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases”.
Use Case 6: Invasive Ductal Carcinoma (IDC) Segmentation
This blog posts explains how to train a deep learning Invasive Ductal Carcinoma (IDC) classifier in accordance with our paper “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases”.
Continue reading Use Case 6: Invasive Ductal Carcinoma (IDC) Segmentation
Use Case 3: Tubule Segmentation
This blog posts explains how to train a deep learning tubule segmentation classifier in accordance with our paper “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases”.